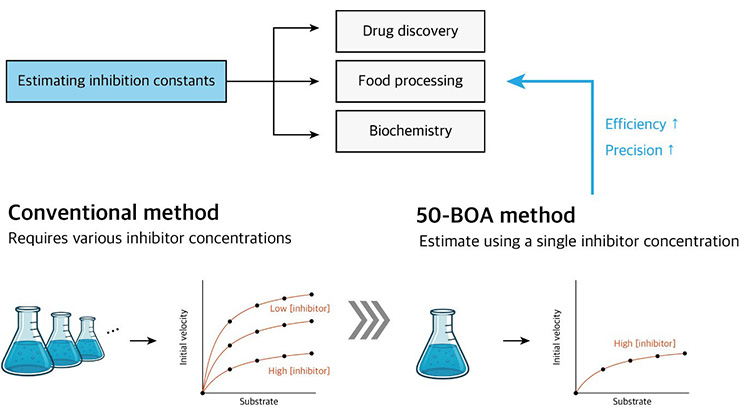
A Model-informed Shortcut to Estimate Inhibition Constants
Inhibition constants are fundamental parameters that quantify drug-drug interactions and guide medication dosing. Conventional estimation methods—currently used in more than 60,000 studies—require extensive experimental data from varied conditions to estimate these constants. We developed a novel approach that substantially reduces the necessary experimental requirements for estimation while preserving, and even sometimes improving, accuracy and precision relative to standard techniques.
The Problem With Conventional Estimation
When a patient takes two drugs simultaneously, one drug (inhibitor) may bind to the target enzyme of another drug (substrate) and interfere with the substrate’s metabolic process (see Figure 1). This phenomenon—known as enzyme inhibition—plays a critical role in clinical pharmacology, as it can lead to increased dosage levels, reduced efficacy, or heightened toxicity of the affected drug [3]. Given inhibitor concentration \(I_T\), the initial metabolic velocity (\(V_0\)) of a substrate with initial concentration \(S_T\) decreases by following the subsequent inhibition model \((1)\):
\[V_0:=\frac{dP}{dt}\Bigg |_{t=0}=\frac{V_\textrm{max}S_T}{K_M(1+\frac{I_T}{K_{ic}})+S_T(1+\frac{I_T}{K_{iu}})}. \tag1\]
Here, \(V_\textrm{max}\) and \(K_M\) are parameters that pertain to the substrate and are usually known in advance from \(I_T=0\) data. \(V_\textrm{max}\) is a maximal metabolic velocity and \(K_M\) is a substrate concentration that reaches half of \(V_\textrm{max}\). Additionally, \(K_{ic}\) and \(K_{iu}\) are the inhibition constants, which respectively serve as dissociation constants of the inhibitor to the enzyme and enzyme-substrate complex (see Figure 2). They not only represent the potency of inhibition but also allow us to infer the mechanism of inhibitors or influence drug dose adjustments. If \(K_{ic}<K_{iu},\) for instance, dose adjustments can help avoid clinical risks; however, doing so is challenging when \(K_{iu}<K_{ic}.\) The precise and accurate estimation of these parameters is therefore critical [3, 8].
![<strong>Figure 2.</strong> Diagram of enzyme inhibition. Inhibitors delay or suppress conversion of a substrate to a product by binding to either the enzyme or enzyme-substrate complex, thereby hindering the catalytic process. Figure courtesy of [6].](/media/dd4dfyfg/figure2.jpg)
Conventionally, scientists estimate inhibition constants by first determining the half maximal inhibitory concentration (\(\textrm{IC}_{50}\)),1 then fitting \((1)\) to initial velocity data from various combinations of \(S_T\) and \(I_T\) around \(\textrm{IC}_{50}\) (see Figure 3) [2]. Although this method has been the standard for decades, its optimality and sufficiency for precise estimation remain unproven. In fact, multiple studies have observed inconsistencies in reported inhibition constants [7, 9].
![<strong>Figure 3.</strong> The conventional approach for estimating inhibition constants consists of three steps: (i) Determine \(\textrm{IC}_{50}\) from experiments with a single \(S_T\) and various \(I_T\). Here, percentage of control activity refers to the initial velocity in the presence of the inhibitor as a percentage of the rate without the inhibitor. (ii) Measure initial velocities across a range of substrate concentrations from low to high for \(K_M\), and across a range of inhibitor concentrations from low to high for \(\textrm{IC}_{50}\). (iii) Estimate inhibition constants by fitting \((1)\) to the collected data. Figure courtesy of [6].](/media/u1iarw3s/figure3.jpg)
A Model-guided Approach
To address the aforementioned issues with conventional estimation methods, we conducted error landscape and sensitivity analyses across various experimental conditions [6]. Our results indicate that data obtained via a single inhibitor concentration \(I_T\) that is greater than both inhibition constants is sufficient for precise estimation (see Figure 4b). In contrast, estimation is highly sensitive to measurement errors when we collect data at \(I_T\) values that are below both inhibition constants (see Figure 4a). These findings suggest that the conventional approach may collect unnecessary or even detrimental data, which can reduce estimation accuracy and precision. But since the true values of the inhibition constants are not known a priori, it is difficult to identify the experimental data in advance that are informative, redundant, or potentially harmful.
![<strong>Figure 4.</strong>Comparison of inhibition constant estimation results via both low inhibitor concentration and sufficiently high concentration. <strong>4a.</strong> During low concentration, the range with low estimation error is too broad and yields inhibition constant values that differ up to 100 times from the actual value. This discrepancy makes it difficult to accurately estimate the inhibition constants. <strong>4b.</strong> In contrast, the use of sufficiently high concentrations narrows the range of low estimation error and allows for more accurate estimation of the inhibition constants. Figure adapted from [6].](/media/ywwfvdxm/figure4.jpg)
IC50 as a Guide
To identify informative experimental data without prior knowledge of the inhibition constants, we focused on \(\textrm{IC}_{50}\) — which scientists typically disregard after initial experimental setup (see Figure 3). \(\textrm{IC}_{50}\) satisfies the well-known weighted harmonic mean relationship with the inhibition constants [5]:
\[\frac{1}{\textrm{IC}_{50}}=\frac{\alpha}{K_{ic}}+\frac{1-\alpha}{K_{iu}}=:\frac{1}{H(K_{ic},K_{iu})}, \qquad \alpha=\frac{K_M}{S_T+K_M}. \tag2\]
This relationship implies that choosing \(I_T\ge\textrm{IC}_{50}\) ensures that either \(I_T\ge K_{ic}\) or \(I_T \ge K_{iu},\) providing a practical criterion for the collection of only informative data while avoiding the bias and sensitivity issues that arise when \(I_T\le\min\{K_{ic},K_{iu}\}.\) We incorporated this relationship into our estimation process as an \({IC}_{50}\)-based regularization term in \((3)\):
\[\textrm{Total error}=\textrm{fitting error}+\lambda \times \Bigg(\frac{\textrm{IC}_{50}-H(K_{ic},K_{iu})}{\textrm{IC}_{50}}\Bigg)^2. \tag3\]
Here, we use the mean of the squared relative error between \(V_0\) data and \((1)\) to calculate fitting error. \(\textrm{IC}_{50}\) is an observed value, \(H(K_{ic},K_{iu})\) is a computed \(\textrm{IC}_{50}\) via \((2)\), and \(\lambda\) is a regularization constant that is obtained with cross-validation.
![<strong>Figure 5.</strong> Incorporation of the \(\textrm{IC}_{50}\)-based optimal approach (50-BOA) can enhance estimation precision. <strong>5a.</strong> When true values \(K_{ic}=1\mu M\) and \(K_{iu}=10\mu M,\) data fitting that uses \(I_T=\textrm{IC}_{50}\) cannot sufficiently achieve precise estimation for \(K_{iu},\) thus indicating a narrower low error region. Incorporation of the regularization error into the fitting error (total error) allows for precise estimation of inhibition constants. <strong>5b.</strong> When two inhibition constants have comparable values, the addition of 50-BOA also enhances the precision. Figure adapted from [6].](/media/u30iu3po/figure5.jpg)
This approach enables precise, accurate estimation of the inhibition constants by using data from a single \(I_T \ge \textrm{IC}_{50},\) even when \(I_T\) is less than both constants (see Figure 5). We combine these strategies and propose the \(IC_{\textit{50}}\)-based optimal approach (50-BOA) for efficient estimation of inhibition constants (see Figure 1).
![<strong>Figure 6.</strong> Comparison of inhibition constant estimation with the conventional method versus the \(\textrm{IC}_{50}\)-based optimal approach (50-BOA) with data from real drug substrate triazolam, inhibitor ketoconazole, and enzyme cytochrome P450 3A4 [4]. Dots and asterisks respectively represent the estimated value of inhibition constants from the conventional method and 50-BOA. The error bars indicate 95 percent confidence intervals (CIs). <strong>6a and 6b.</strong> 50-BOA demonstrates an accuracy level that is analogous to the conventional method, with CIs that were either similar or narrower. <strong>6c.</strong> The number of experiments is reduced by a factor of \(1/7\) when compared to the conventional method. Figure courtesy of the authors.](/media/ywtcn3hf/figure6.jpg)
Validating 50-BOA: Accurate and Efficient Estimation With Real Data
Applying 50-BOA to experimental data achieves an accuracy level that is comparable to the conventional approach (see Figures 6a and 6b). Additionally, the 95 percent confidence intervals are similar to or narrower than those from the conventional method. As a result, 50-BOA allows for equal or improved accuracy and precision while reducing the required experimental data by more than 75 percent (see Figure 6c).
We expect our study to enhance the efficiency of enzyme inhibition evaluation in both drug development and other relevant fields like the food industry, which utilizes enzyme inhibition to prevent food browning and spoilage [1]. Moreover, we anticipate that our work will catalyze future research into optimal, experimentally-feasible designs — ultimately extending beyond enzyme inhibition to various biological systems.
1 Half maximal inhibitory concentration (\(\textrm{IC}_{50}\)) is an inhibitor concentration where the effect of inhibition compared to \(I_T=0\) reaches 50 percent.
Acknowledgments: We would like to acknowledge the coauthors of this study: Sang Kyum Kim, Hwi-yeol Yun, and Jang Su Jeon of Chungnam National University.
References
[1] Ackaah‐Gyasi, N.A., Zhang, Y., & Simpson, B.K. (2016). Enzymes inhibitors: Food and non‐food impacts. In R. Rai V. (Ed.), Advances in food biology (pp. 191-206). Hoboken, NJ: Wiley.
[2] Copeland, R.A. (2013). Evaluation of enzyme inhibitors in drug discovery: A guide for medicinal chemists and pharmacologists. Hoboken, NJ: John Wiley & Sons.
[3] Deodhar, M., Al Rihani, S.B., Arwood, M.J., Darakjian, L., Dow, P., Turgeon, J., & Michaud, V. (2020). Mechanisms of CYP450 inhibition: Understanding drug-drug interactions due to mechanism-based inhibition in clinical practice. Pharmaceutics, 12(9), 846.
[4] Greenblatt, D.J., Zhao, Y., Venkatakrishnan, K., Duan, S.X., Harmatz, J.S., Parent, S.J., … von Moltke, L.L. (2011). Mechanism of cytochrome P450-3A inhibition by ketoconazole. J. Pharm. Pharmacol., 63(2), 214-221.
[5] Haupt, L.J., Kazmi, F., Ogilvie, B.W., Buckley, D.B., Smith, B.D., Leatherman, S., … Parkinson, A. (2015). The reliability of estimating Ki values for direct, reversible inhibition of cytochrome P450 enzymes from corresponding IC50 values: A retrospective analysis of 343 experiments. Drug Metab. Dispos., 43(11), 1744-1750.
[6] Jang, H.J., Song, Y.M., Jeon, J.S., Yun, H.-y., Kim, S.K., & Kim, J.K. (2025). Optimizing enzyme inhibition analysis: Precise estimation with a single inhibitor concentration. Nat. Commun., 16(1), 5217.
[7] McFeely, S.J., Ritchie, T.K., & Ragueneau‐Majlessi, I. (2020). Variability in in vitro OATP1B1/1B3 inhibition data: Impact of incubation conditions on variability and subsequent drug interaction predictions. Clin. Transl. Sci., 13(1), 47-52.
[8] Palleria, C., Di Paolo, A., Giofrè, C., Caglioti, C., Leuzzi, G., Siniscalchi, A., … Gallelli, L. (2013). Pharmacokinetic drug-drug interaction and their implication in clinical management. J. Res. Med. Sci., 18(7), 601-610.
[9] Quinney, S.K., Knopp, S., Chang, C., Hall, S.D., & Li, L. (2013). Integration of in vitro binding mechanism into the semiphysiologically based pharmacokinetic interaction model between ketoconazole and midazolam. CPT Pharmacometrics Syst. Pharmacol., 2(9), e75.
About the Authors
Hyeong Jun Jang
Undergraduate student, Korea Advanced Institute of Science and Technology
Hyeong Jun Jang is an undergraduate student in the School of Transdisciplinary Studies at the Korea Advanced Institute of Science and Technology. His research focuses on the mathematical model-based analysis and optimization of experimental setups in pharmacokinetics.
Yun Min Song
Postdoctoral fellow, Institute for Basic Science
Yun Min Song is a postdoctoral fellow in the Biomedical Mathematics Group at the Institute for Basic Science in South Korea. He holds a Ph.D. in mathematical sciences from the Korea Advanced Institute of Science and Technology. Song’s work integrates mathematical modeling, statistical inference, and artificial intelligence to address complex biomedical problems, including pharmacokinetics and the analysis of wearable-device data.

Jae Kyoung Kim
Associate professor, Korea Advanced Institute of Science and Technology
Jae Kyoung Kim is an associate professor in the Department of Mathematical Sciences at the Korea Advanced Institute of Science and Technology and chief investigator in the Biomedical Mathematics Group at the Institute for Basic Science in South Korea. He holds a Ph.D. in applied and interdisciplinary mathematics from the University of Michigan. Kim’s research combines nonlinear dynamics, stochastic processes, and scientific computing to address critical biological and medical problems, including sleep disorders.