Networked Epidemiology for COVID-19
Computational epidemiology aims to develop computer models and decision support systems that understand, predict, and control a disease’s spatiotemporal diffusion throughout a population. Researchers can use these models to forecast an epidemic’s future course, allocate scarce resources and assess depletion of current resources, infer disease parameters, and evaluate various interventions. Individual behavior and public policy are critical in understanding and controlling infectious diseases, and computational techniques provide a potentially powerful study tool. The COVID-19 pandemic has had significant social, health, economic, and political ramifications worldwide, and its impact will undoubtedly continue to grow in the coming months. Here we outline an approach to support the COVID-19 response with examples that are rooted in network science and data-driven modeling.
Computational Models: From ODEs to Multi-scale Networks
Compartmental mass action models are a cornerstone of mathematical epidemiology. They partition a homogeneous population into a small set of compartments that represent the possible disease states—e.g., susceptible \((S)\), infectious \((I)\), and removed \((R)\)—and specify transition rates between states. Epidemiologists have successfully used these models in the past and continue to do so. Desirable features of compartmental models include their analytical tractability—one can analyze simple dynamic models using scalable numerical simulations, or solve them to yield closed-form solutions or asymptotic limits––and their light demands on computational resources.
An alternative way to study epidemics involves explicit representation of the underlying contact structure that drives them [1-3]. We focus on networked models that consider epidemic spread on an undirected social interaction network \(G(V,E)\) over population \(V\). Each edge \(e=(u,v) \in E\) implies that individuals (also referred to as nodes) \(u, v \in V\) interact. The specific form of interaction depends on the disease in question; for example, sexually transmitted diseases require physical sexual contact, while respiratory illnesses necessitate only physical proximity. Let \(N(v)\) denote the set of neighbors of \(v\). The SIR model on graph \(G\) is a dynamical process during which each node is in either an \(S\), \(I\), or \(R\) state. Infection can potentially spread from \(u\) to \(v\) along edge \(e=(u,v)\) with a probability of \(\beta(e,t)\) at time \(t\) after \(u\) becomes infected, which is conditional on node \(v\) remaining uninfected until time \(t\); this is a discrete version of the infection rate for the aforementioned ordinary differential equation (ODE) model. We allow \(I(t)\) to denote the set of nodes that become infected at time \(t\). The random subset of edges on which the infections spread represents a disease outcome and is called a dendrogram. This dynamical system begins with a configuration that features one or more nodes in state \(I\) and ultimately reaches a fixed point where all nodes are in states \(S\) or \(R\). Some key topics of interest are as follows: (i) Characterization of aspects of \(I(t)\) (the epicurve)—such as its peak, the time at which the peak occurs, and its integral (the outbreak size)—as a function of the disease model parameters and network structure; and (ii) the effectiveness of various interventions, including vaccination (which can be modeled as node deletions) and social distancing (which can be modeled as edge deletions).
Networks: Scale, Structure, and Detail
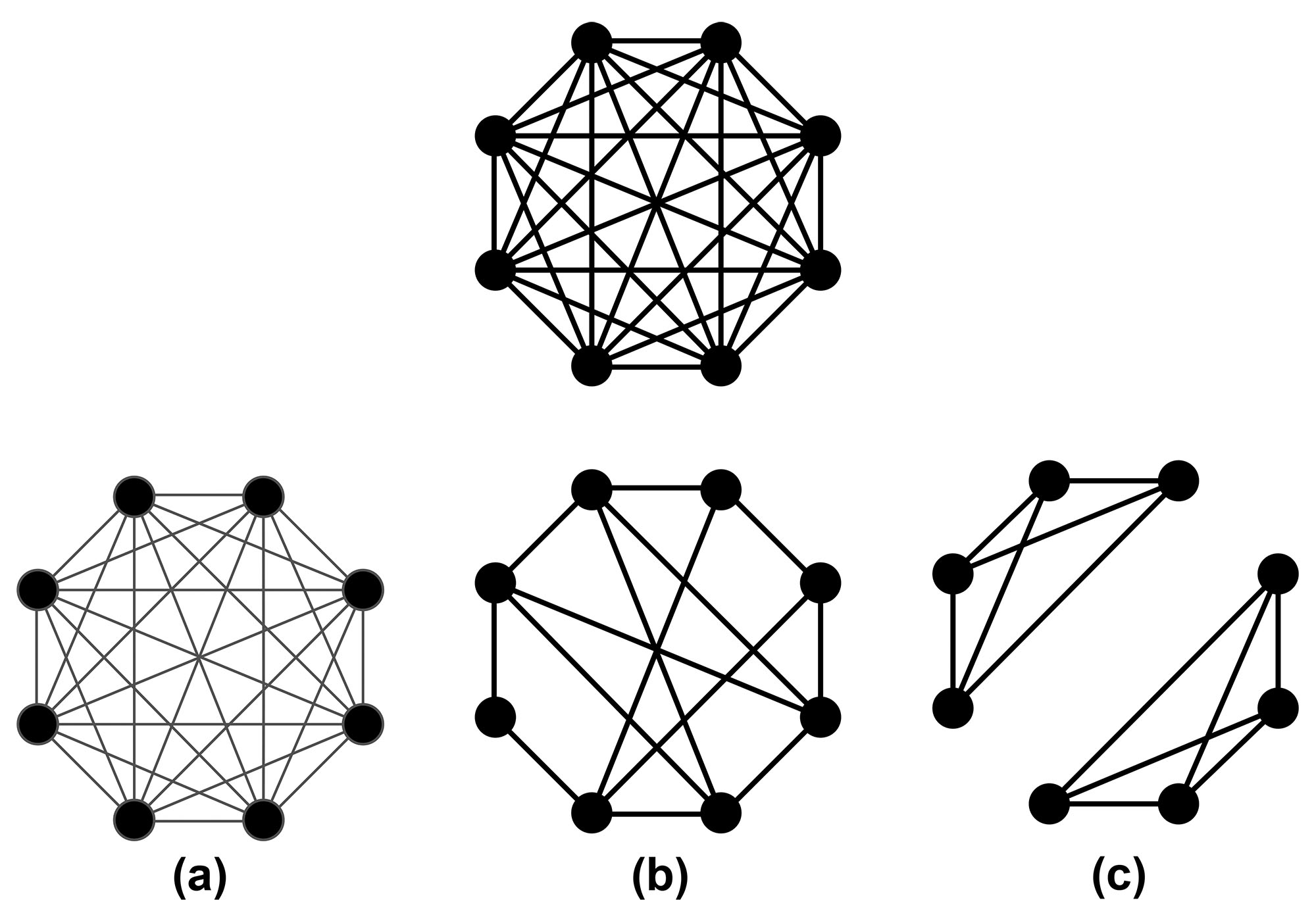
One must treat the network \(G(V,E)\) as a first-class model parameter, on equal footing with disease-related parameters like transmissibility and incubation period. This step simply acknowledges the importance of parameters that are swept under the rug by mass action assumption. Because certain combinations of parameters—such as transmissibility and network structure—are not separately identifiable, problems with mass action assumption are not immediately obvious. Instead, they arise when one attempts to understand the effects of interventions.
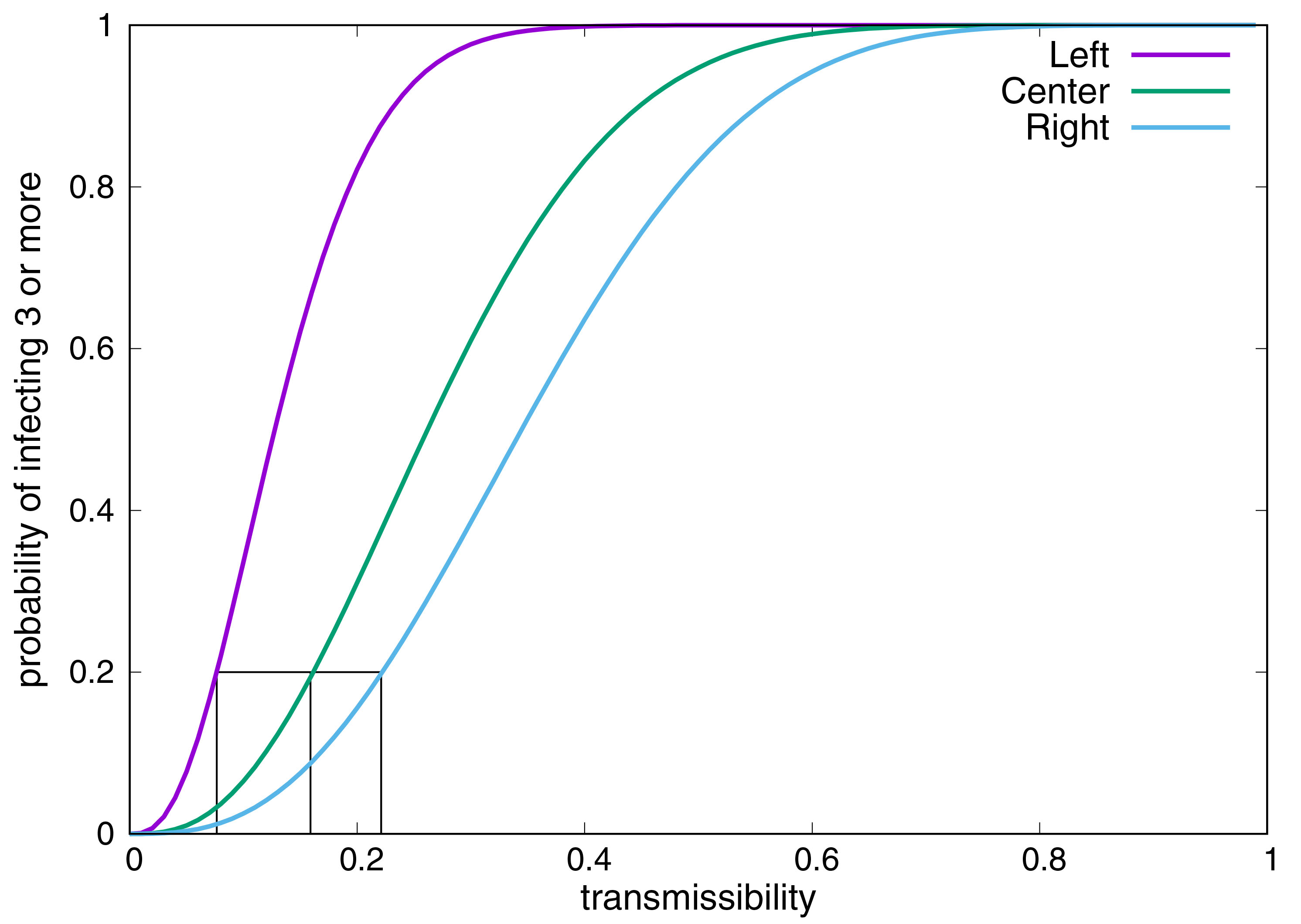
For example, consider the following intervention: reduce contacts by 50 percent. Figure 1 illustrates three plausible interpretations. Figure 1a reduces the probability of transmission (represented by an edge weight) in each contact by 50 percent, 1b reduces the number of contacts by 50 percent, and 1c cuts the graph in half. The dynamics on each of these graphs are clearly different. Figure 2 depicts the probability of a single, randomly selected person infecting as many as three other people on each graph as a function of the probability of transmission. One can calibrate a model’s transmissibility for a range of outbreak sizes to yield the same probability of large outbreaks on each graph: 0.2 in Figure 2. Given only the attack rate, the network structure and transmissibility are thus not separately identifiable.
The only way to represent changes in network structure in mass action models is to adjust transmissibility, but this often has unrealistic consequences for model outcomes (in addition to outbreak size). In particular, a “stay at home” policy is best represented by the network in Figure 1c. A mass action model of staying at home severely distorts the epidemic curve. The true policy impact is given by the sum of many outbreaks in cliques whose sizes are drawn from the distribution of household sizes; in contrast, a mass action model attempts to fit an outbreak among the entire population with an extremely small transmissibility.
Building Contact Networks
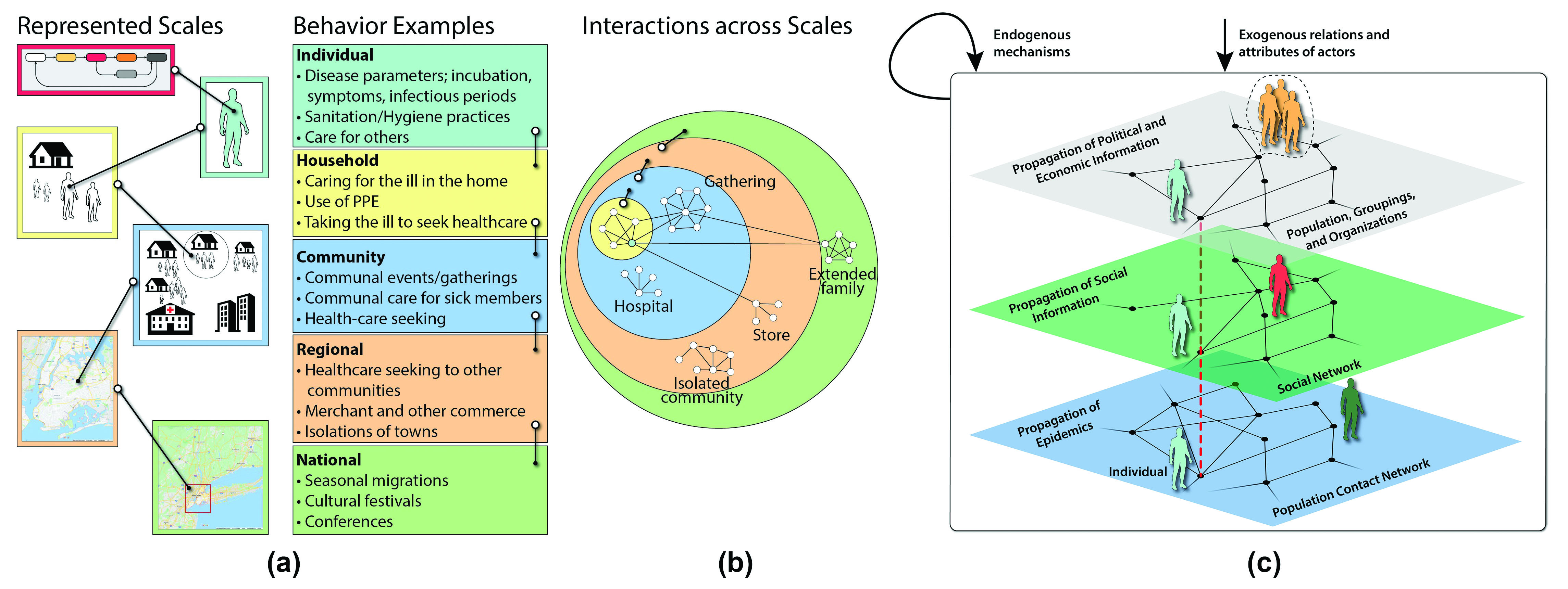
The study of epidemics leads to multi-scale, multi-layer (MSML) networks (see Figure 3). Each network captures different types of interactions and forms the underlying fabric for a distinct contagion process. Constructing social contact networks with sufficient accuracy to model disease spread in cities like Los Angeles or New York City is challenging. Researchers cannot construct such networks by using extensive measurements except in very simple, restricted situations; doing so would require knowledge of every individual’s demographics, activities, and locations, which would be both technologically impossible and ethically questionable. So, how can we accurately represent a city’s populace? One must assemble the networks synthetically by integrating or fusing available datasets with simulation-based generative methods. Unlike simple random graph techniques, these methods synthesize networks by utilizing real-world data sources and combining them with behavioral and social theories.
Scalable Simulations to Study Epidemic Dynamics over Networks
It is difficult to apply an analytical approach in a realistic setting that examines the effects of interventions (which consist of individual and collective behavior at different levels) on epidemics’ dynamics. The complication stems from the unstructured nature of real-world social contact networks, which represent the interactions and heterogeneities between the individuals. Therefore, simulations based on high-performance computing are often the only feasible methods for studying networked epidemic models in large-scale population settings. Our group has developed a variety of these simulation environments to mimic disease spread in large social contact networks.
Network-based Simulations and Algorithmic Workflows during COVID-19 Response
The ongoing COVID-19 pandemic illustrates the importance of MSML networks in representing the underlying interaction structure. Typical studies demonstrate the utility of network models and algorithmic workflows for managing epidemic response across various scales (international, subnational, and community-level) and stages (emergence, containment, and mitigation).
We analyzed global airline traffic to ascertain importation risk for various countries during the early stages of a pandemic. Using data on the number of passengers between international airports, we constructed a flow network on which COVID-19 could potentially spread and reach other countries. This representation—coupled with a disease model at each node—allows us to characterize the time for case emergence, assuming a given origin node. Furthermore, a structural characterization of this network (namely effective distance) captures disease emergence times more successfully than purely geographical distance between airports. Such a network representation—with actual flow volumes labeled by airlines—permitted us to implement interventions like airline suspensions and evaluate the subsequent impact on delaying COVID-19’s arrival in various locations.
Networked epidemiology enables the study of social distancing policies, such as voluntary home isolation and school and workplace closures. Improved testing and social distancing—two current responses to COVID-19—work in tandem and must be studied together. Detailed network models help researchers examine better case isolation and household quarantine (products of increased testing) while simultaneously mimicking the effects of workplace and nonessential business closures in reduced mixing within the population. Similarly, an increased push for privacy-preserving contact tracing efforts via smartphones has emerged in Western countries, following its success in China and South Korea. Individual-level contact networks allow for assimilation of such contact traces (equivalent to the dendrograms of disease spread) and evaluation of the effects of coverage and compliance.
A final example involves determining COVID-19’s burden on healthcare infrastructure and exploring possible mitigation strategies. The healthcare burden depends on many factors, including level of social distancing and regional testing, current healthcare resources, and population demographics. Various social distancing measures are attempting to “flatten the curve” and reduce the overload and potential collapse of healthcare systems due to surges in critical cases. Our initial results suggest that patient transfers can have a tangible effect in reducing the healthcare resource deficit.
Traditional ODE models can effectively guide the development of “rules of thumb” for epidemic response. However, when it comes to assessing particular responses to specific outbreaks, a faithful representation in the form of a networked dynamical system is more useful.
Acknowledgments: The authors would like to thank members of the Network Systems Science and Advanced Computing Division at the University of Virginia (UVA) for useful discussion. This work was partially supported by National Institutes of Health Grant 1R01GM109718, National Science Foundation (NSF) BIG DATA Grant IIS-1633028, NSF DIBBS Grant ACI-1443054, NSF Grant No. OAC-1916805, NSF Expeditions in Computing Grant CCF-1918656 and CCF-1917819, U.S. Centers for Disease Control and Prevention 75D30119C05935, DTRA subcontract/ARA S-D00189-15-TO-01-UVA, and a collaborative seed grant from UVA’s Global Infectious Disease Institute.
References
[1] Eubank, S., Guclu, H., Kumar, V.A., Marathe, M.V., Srinivasan, A., Toroczkai, Z., & Wang, N. (2004). Modelling disease outbreaks in realistic urban social networks. Nature, 429(6988), 180-184.
[2] Halloran, M.E., Ferguson, N.M., Eubank, S., Longini, I.M., Cummings, D.A.T., Lewis, B., ... & Cooley, P. (2008). Modeling targeted layered containment of an influenza pandemic in the United States. Proc. Nat. Acad. Sci., 105(12), 4639-4644.
[3] Marathe, M., & Vullikanti, A.K.S. (2013). Computational epidemiology. Comm. ACM, 56(7), 88-96.Further Reading
Chinazzi, M., Davis, J.T., Ajelli, M., Gioannini, C., Litvinova, M., Merler, S., ..., Vespignani, A. (2020). The effect of travel restrictions on the spread of the 2019 novel coronavirus (COVID-19) outbreak. Science, 368(6489), 395-400.
Ferguson, N., Laydon, D., Nedjati-Gilani, G., Imai, N., Ainslie, K., Baguelin, M., ... & Ghani, A.C. (2020). Report 9: Impact of non-pharmaceutical interventions (NPIs) to reduce COVID19 mortality and healthcare demand. Imperial College London.
Levin, S.A. (Ed.). (1994). Frontiers in mathematical biology. Berlin, Germany: Springer.
Moghadas, S.M., Shoukat, A., Fitzpatrick, M.C., Wells, C.R., Sah, P., Pandey, A., ..., Galvani, A.P. (2020). Projecting hospital utilization during the COVID-19 outbreaks in the United States. Proc. Nat. Acad. Sci., 117(16), 9122-9126.
Perrings, C., Levin, S., & Daszak, P. (2018). The economics of infectious disease, trade and pandemic risk. EcoHealth, 15, 241-243.
Verity, R., Okell, L.C., Dorigatti, I., Winskill, P., Whittaker, C., Imai, N., ... & Ferguson, N.M. (2020). Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect. Dis.
About the Authors
Jiangzhuo Chen
Research Associate Professor, University of Virginia
Jiangzhuo Chen is a research associate professor in the Network Systems Science and Advanced Computing Division of the University of Virginia’s Biocomplexity Institute.
Simon A. Levin
James S. McDonnell Distinguished University Professor, Princeton University
Simon A. Levin is the James S. McDonnell Distinguished University Professor in Ecology and Evolutionary Biology at Princeton University. His research interests are in theoretical and applied ecology, especially the interface with socio-economic systems. Levin is a SIAM Fellow and a 2014 National Medal of Science recipient.
Stephen Eubank
Professor, University of Virginia
Stephen Eubank is a professor in the Department of Public Health Sciences and the Biocomplexity Institute at the University of Virginia.
Henning Mortveit
Associate Professor, University of Virginia
Henning Mortveit is an associate professor in the Department of Engineering Systems and Environment and the Biocomplexity Institute at the University of Virginia.
Srinivasan Venkatramanan
Research Assistant Professor, Biocomplexity Institute
Srinivasan Venkatramanan is a research assistant professor at the Network Systems Science and Advanced Computing division of the University of Virginia’s Biocomplexity Institute. He develops, analyzes, and optimizes computational models for complex systems that arise in the domains of epidemiology and food security.
Anil Vullikanti
Professor, University of Virginia
Anil Vullikanti is a professor in the Department of Computer Science and the Biocomplexity Institute at the University of Virginia.
Madhav Marathe
Distinguished professor, Biocomplexity Institute
Madhav Marathe is an endowed Distinguished Professor in Biocomplexity, director of the Network Systems Science and Advanced Computing Division in the Biocomplexity Institute, and a tenured professor of computer science at the University of Virginia. His areas of expertise are network science, artificial intelligence, high-performance computing, computational epidemiology, biological and socially coupled systems, and data analytics.
Stay Up-to-Date with Email Alerts
Sign up for our monthly newsletter and emails about other topics of your choosing.



