Novel Hyperspectral Imaging-based Method Enhances Segmentation of Ocular Tissue
Hyperspectral imaging (HSI) is an advanced technique that combines spectroscopy and digital imaging to capture detailed information about an object over many continuous wavelengths both within and beyond the visible spectrum. This powerful, noninvasive methodology seeks to attain the spectrum for each pixel of the image in question. Whereas RGB images have only three channels (red, green, and blue), HSI images are three-dimensional (3D) hypercubes; just one pixel from this hypercube can reveal a continuous spectral signature.
HSI allows scientists to locate particular objects, recognize certain materials, and detect specific patterns. It finds applications across a wide range of settings, including surveillance and security, historical conservation, food safety and control, environmental analysis and agriculture, and medicine. In the medical field, biomedical HSI (BHSI) is an emerging imaging modality for biomedical applications that provides diagnostic information about tissue physiology, morphology, and composition; it is especially relevant to disease diagnosis and image-guided surgery. BHSI works by (i) imaging samples from a hyperspectral camera, (ii) processing the images for training algorithms, and (iii) applying the trained algorithms for detection (see Figure 1).
![<strong>Figure 1.</strong> Workflow for hyperspectral imaging and training. Figure courtesy of [1].](/media/vpwm5xk3/figure1.jpg)
During a minisymposium presentation at the Third Joint SIAM/CAIMS Annual Meetings, which are currently taking place in Montréal, Québec, Canada, Na Yu of Toronto Metropolitan University utilized BHSI and machine learning techniques to improve the segmentation and analysis of hyperspectral images of eye tissue. She and her team obtained their BHSI data from the Eye Pathology Laboratory at St. Michael’s Hospital in Toronto. “Our BHSI images are taken by an imaging mapping spectrometer with a custom-modified camera that can capture reflected fluorescent light near the infrared spectrum,” Yu said. “The produced hypercubes are layered images taken across 78 different wavelengths, each four nanometers (nm) apart and ranging from 528 nm to 836 nm.”
Yu’s study focuses on the four main tissue sections of the eye: the retina (both photoreceptors and the inner layer), choroid, sclera, and muscle. All of these ocular tissues exhibit autofluorescence, which means that they naturally emit light when exposed to certain wavelengths. Yu hopes that segmenting HSI images into these key tissue types will allow practitioners to precisely identify and differentiate between tissues and conditions, thus enhancing the early detection and continuous monitoring of eye diseases.
However, several challenges inhibit her efforts. Given the limited number of available BHSI images—especially for sections of the eye—a restrictively small sample size is common, as is a lack of labeled BHSI imagery. High dimensionality, noise, and imaging artifacts are also present in BHSI data. “All of these vectors highlight the need for efficient and accurate image processing techniques, especially segmentation,” Yu said.
Next, Yu introduced a well-known tool called UNET: a U-shaped convolutional neural network for biomedical image segmentation. Developed in 2015, UNET maintains its popularity in the present day due to its consistently high performance, even with a limited number of training samples. It comprises two parts: (i) An encoder network that acts as the feature extractor, learning an abstract representation of the input image through a sequence of four encoder blocks; and (ii) a decoder network that uses the abstract representation to generate a semantic segmentation map.
Despite its proficiency, the classic UNET is not appropriate for Yu’s needs because it is meant for two-dimensional data, whereas BHSI generates a 3D hypercube. And while a 3D-UNET that handles 3D RGB images does exist, it is likewise unsuitable in this context. Instead, Yu proposed a novel method that relies on data augmentation: the artificial generation of new data from existing data to train new machine learning methods. This process involves the spatial transformation of data via rotations and flips and yields a totally trained set of 1,020 items, a validation set of 306 items, and a test set of 132 items.
The subsequent step involves dimension reduction for the high-dimensional hypercube. Using principal component analysis, Yu found that reduction to between three and five principal components retains 99.5 percent of the variance in the original hypercube; it also speeds up the training process. She calls her resulting architecture the 3D-R2A2-UNet++ Architecture. It is quite complex, so Yu highlighted just two components: (i) The convolutional block attention module for boundary detection and (ii) the dual focus gate, which processes spatial and spectral information and accelerates convergence of the UNET algorithm.
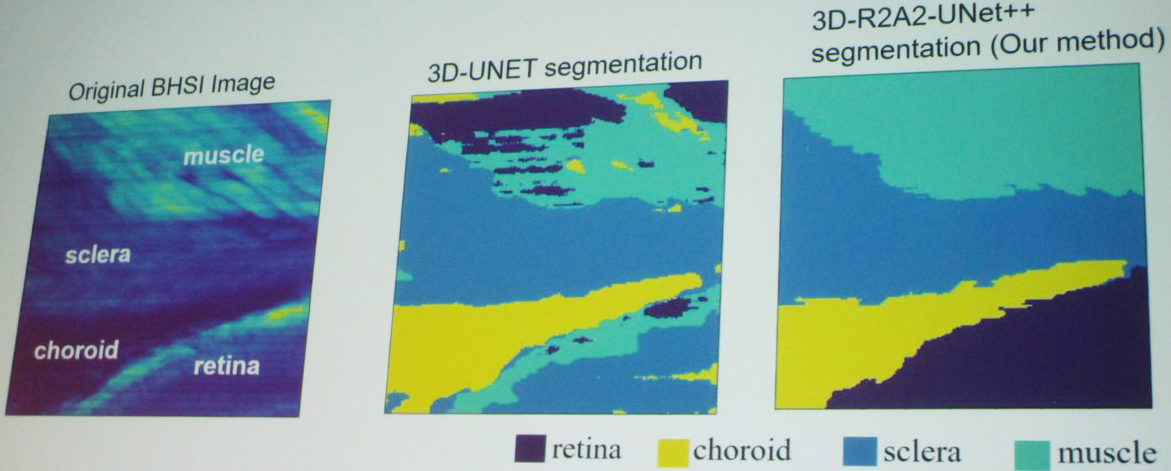
Yu then shared a visualization of her segmentation results, which compared the original BHSI image with the 3D-UNET segmentation and her own 3D-R2A2-UNet++ segmentation (see Figure 2). “By visual comparison, you can see that our method is much better than the standard 3D network,” she said. A corresponding quantitative comparison used five performance metrics—dice score, accuracy, recall, precision, and mean Intersection over Union—to corroborate these visual results.
Ultimately, Yu’s unique BHSI method and subsequent incorporation of machine learning enhances the segmentation of ocular tissue. These collective advancements have the potential to enable early detection of eye abnormalities and ultimately advance the use of BHSI in future health studies.
References
[1] Khouj, Y., Dawson, J., Coad, J., & Vona-Davis, L. (2018). Hyperspectral imaging and k-means classification for histologic evaluation of ductal carcinoma in situ. Front. Oncol., 8, 17.
About the Author
Lina Sorg
Managing editor, SIAM News
Lina Sorg is the managing editor of SIAM News.

Stay Up-to-Date with Email Alerts
Sign up for our monthly newsletter and emails about other topics of your choosing.